 Summary
Summary
In this article, I'll describe fluorescence observations of the ciliate Climacostomum virens, that I have in culture.
Method
Climacostomum virens is a heterotrich ciliate species (phylum: Ciliophora). This species typically carries symbiotic zoochlorelles, that cause the green color of the specimen. In a forgotten sample I found this species months after taking the sample from nature. As the sample was kept in the dark, the specimen carried only few zoochlorelles at that time, thus appeared more transparent. Their main food consisted of Bacillariophyta (diatoms) at that time. Over months I failed to switch the ciliate diet to green alga. In January 2021, I found one of my cultures working. This is good new, as it enables long-term studies of this ciliate in my laboratory.
The fluorescence staing shown here is another example of my double staining method using Ho342 and acridine orange (AO), developed to determine and observe ciliate species. AO stains acidic organelles, that are mainly mucocysts and acidic organelles carrying digesting substances. With Climacostomum virens the staining results are mainly showing the cortical granules, organells along the adoral zone of membranelles and inner phagosomes digesting the food. The macronucleus is stained in clear blue from Ho432. Cortical granules of Climasostomum are defensive organelles that produce toxic substances. These toxines have been recently analysed and were named after the species "climacostol". These small organelles can be detected at high resultion with a good light microscope. Correlative observations show, that these are the same organelles, stained by AO. Also stained are organelles along the ingesting tract from the perisom with its adoral zone of membranelles and deeper to the end, where new phagosomes are building up and later digest the food. As we cannot distinguish between the cortical granules and those surrounding the persistome channel, at least these organelles appear all acidic. It is likely these organelles along the digestive tract may also carry toxic substances, as Climacostomum virens is not only defensive, but also a large predator.
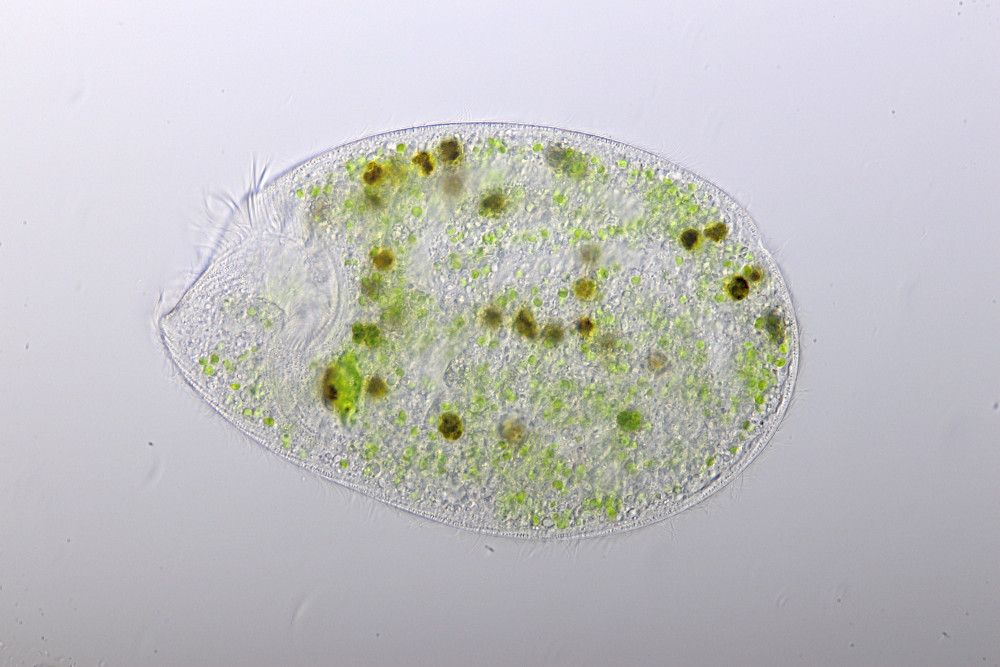
Image 1: Bright field image of Climacostomum virens. The long s-shaped peristome can be clearly seen. Small green algae are found in the main body along with larger phagosomes digesting Euglena, provided as food source.
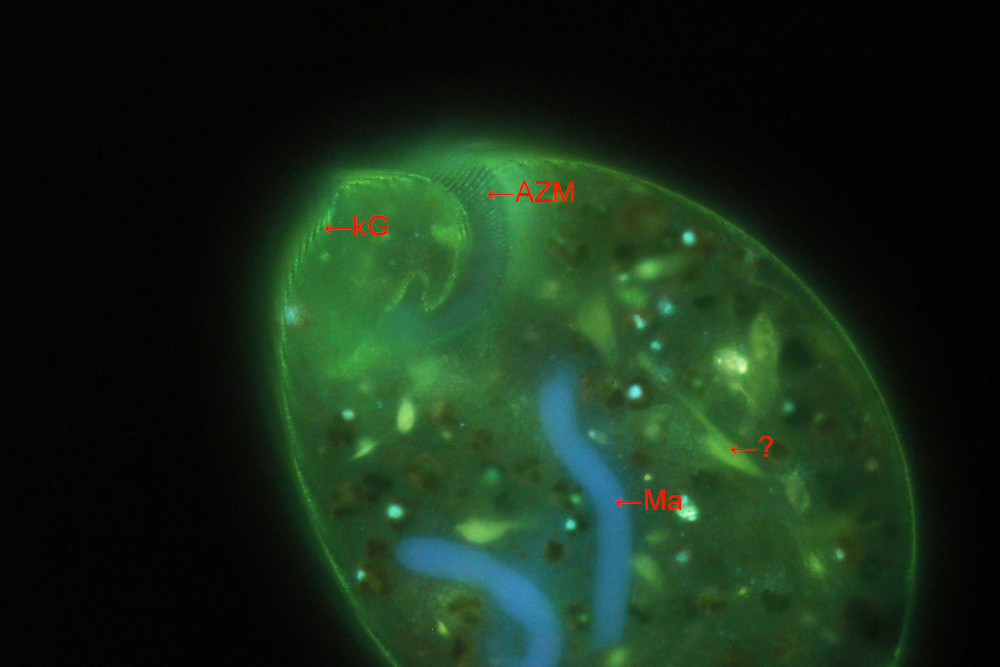
Image 2: Fluorescence image of a living cell with details of the peristome and adoral zone of membranelles (AZM) and cortical rows of acidic organelles (kG). The large macronucleus (Ma) is stained in blue. A question mark denotes inner fibers stained in green. The nature of these fibres is yet unknown to me.
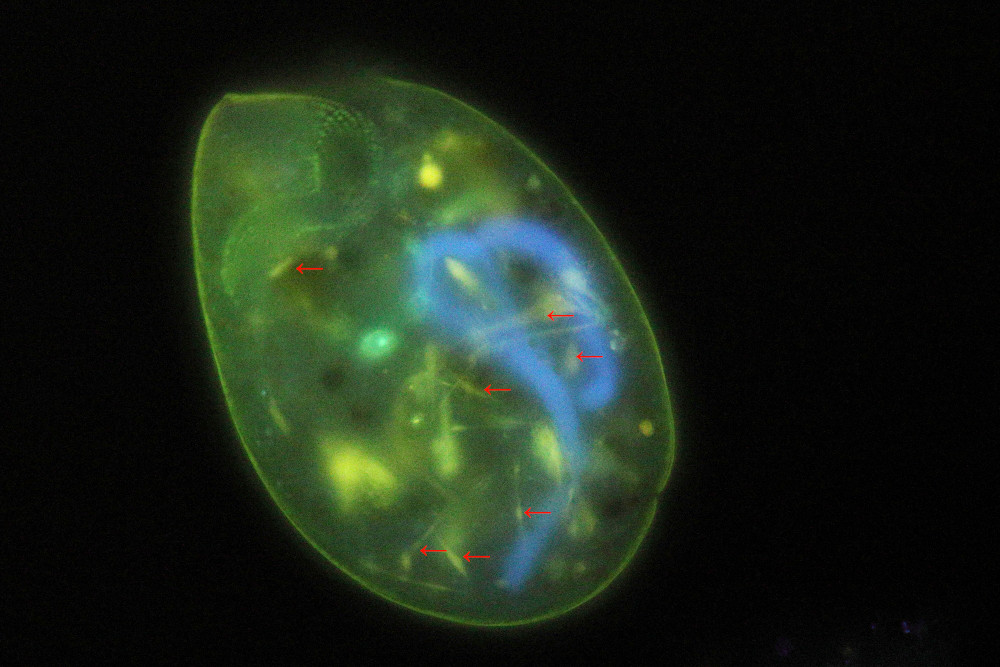
Image 3: Another full size image of the same specimen shown in image 2.
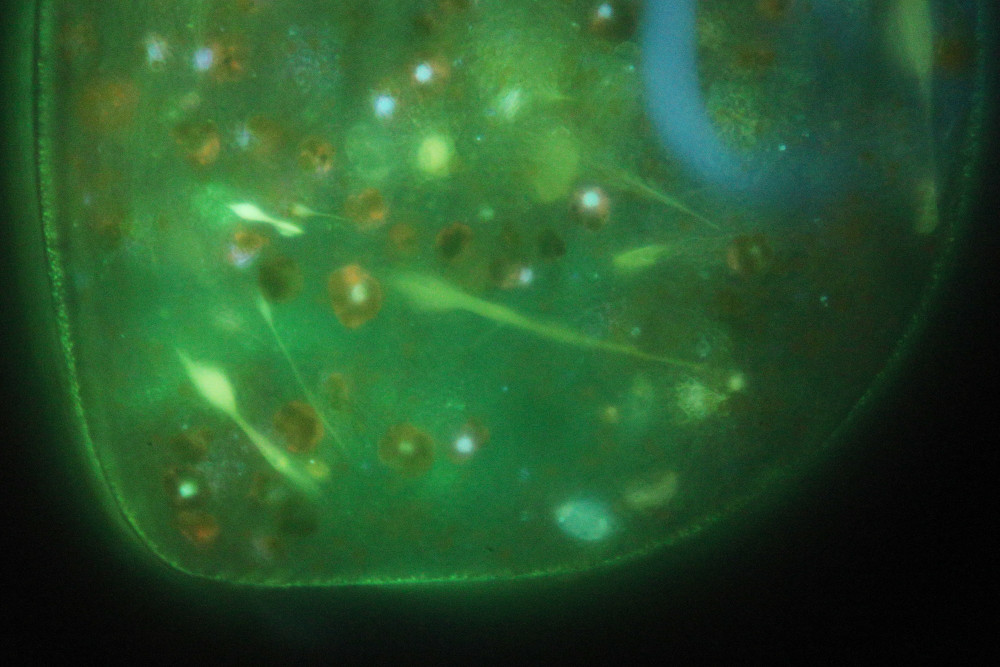
Bild 4: Fluorescence image at highest magnification showing some fibres. Green symbiotic algae appear in dark red with some nuclei stained in blue. The triple-band fluorescence filter cube suppresses well auto-fluorescence of chlorophyll. Otherwise, the bright red color from chlorophyll auto-fluorescence could be the dominant image intensity and other details might be impossible to study.
Observational data of photomicrographs
Microscope and camera: Zeiss Axio Lab.A1, Canon EOS 77D. Brightfield image taken in optimized oblique illumination. Image 1-3 taken with a water immersion optics of type Zeiss C-Apochromat 40x/1,2 W. Image 4 was taken using a Zeiss LCI Plan-Neofluar 63x/1,3 DIC in water immersion. Fluorescence excitation at 385 nm using a AHF triple-band filter cube of type F66-412.
Literature
Masaki, M. E. et al., 2004. Climacostol, a defense toxin of Climacostomum virens (protozoa, ciliata), and its congeners. Tetrahedron, 60(33), 7041-7048.
Bauer, T., 2019. Fluoreszenz-Doppelfärbung mit Hoechst 33342 und Acridinorange zur Bestimmung der Arten von Ciliaten (Phylum: Ciliophora). Mikroskopie; 1: 2-19.